What is a Generalized Linear Model (GLM)?
Generalized Linear Model (GLM) is a statistical model that can be applied even when the objective variable follows a probability distribution other than the normal distribution.
Simple or multiple regression analysis is called a linear model and is a statistical model that assumes that the objective variable follows a normal distribution. If the objective variable cannot be assumed to follow a normal distribution, for example, if the objective variable is a discrete variable such as categorical or count data, the linear model is not applicable. GLM can handle not only the normal distribution but also other distributions called exponential distribution families such as binomial, gamma, and Poisson distributions.
In the GLM, the relationship between the objective and explanatory variables need not be a simple linear equation, but can be expressed as follows:
Combination of probability distribution and link function
Commonly used combinations of probability distributions and link functions are as below table.
| Probability distribution | Link function | |
|---|---|---|
| Discrete | Binomial distribution | logit |
| Discrete | Poisson distribution | log |
| Discrete | Negative binomial distribution | log |
| Continuous | Gamma distribution | log |
| Continuous | Normal distribution | identity |
Constructing the GLM
Let us actually construct the GLM. Let us consider the seed count data of 100 fictitious plants. The data consists of the following columns:
- y: Number of seeds of the individuals
- x: Size of individuals
- f: whether the individuals have been treated with fertilizer or not (C: without fertilizer, T: with fertilizer)
import pandas as pd
# number of seeds
y = [6,6,6,12,10,4,9,9,9,11,6,10,6,10,11,8,3,8,5,5,4,11,5,10,6,6,7,9,3,10,2,9,10,
5,11,10,4,8,9,12,8,9,8,6,6,10,10,9,12,6,14,6,7,9,6,7,9,13,9,13,7,8,10,7,12,
6,15,3,4,6,10,8,8,7,5,6,8,9,9,6,7,10,6,11,11,11,5,6,4,5,6,5,8,5,9,8,6,8,7,9]
# body size
x = [8.31,9.44,9.5,9.07,10.16,8.32,10.61,10.06,9.93,10.43,10.36,10.15,10.92,8.85,
9.42,11.11,8.02,11.93,8.55,7.19,9.83,10.79,8.89,10.09,11.63,10.21,9.45,10.44,
9.44,10.48,9.43,10.32,10.33,8.5,9.41,8.96,9.67,10.26,10.36,11.8,10.94,10.25,
8.74,10.46,9.37,9.74,8.95,8.74,11.32,9.25,10.14,9.05,9.89,8.76,12.04,9.91,
9.84,11.87,10.16,9.34,10.17,10.99,9.19,10.67,10.96,10.55,9.69,10.91,9.6,
12.37,10.54,11.3,12.4,10.18,9.53,10.24,11.76,9.52,10.4,9.96,10.3,11.54,9.42,
11.28,9.73,10.78,10.21,10.51,10.73,8.85,11.2,9.86,11.54,10.03,11.88,9.15,8.52,
10.24,10.86,9.97]
# feed (C: control, T: treatment)
f = ['C','C','C','C','C','C','C','C','C','C','C','C','C','C','C','C','C','C','C',
'C','C','C','C','C','C','C','C','C','C','C','C','C','C','C','C','C','C','C',
'C','C','C','C','C','C','C','C','C','C','C','C','T','T','T','T','T','T','T',
'T','T','T','T','T','T','T','T','T','T','T','T','T','T','T','T','T','T','T',
'T','T','T','T','T','T','T','T','T','T','T','T','T','T','T','T','T','T','T',
'T','T','T','T','T']
df = pd.DataFrame(list(zip(y, x, f)), columns =['y', 'x', 'f'])
display(df.head())
| y | x | f | |
|---|---|---|---|
| 0 | 6 | 8.31 | C |
| 1 | 6 | 9.44 | C |
| 2 | 6 | 9.50 | C |
| 3 | 12 | 9.07 | C |
| 4 | 10 | 10.16 | C |
Data investigation
Let us check descriptive statistics.
df.describe()
| y | x | |
|---|---|---|
| count | 100.000000 | 100.000000 |
| mean | 7.830000 | 10.089100 |
| std | 2.624881 | 1.008049 |
| min | 2.000000 | 7.190000 |
| 25% | 6.000000 | 9.427500 |
| 50% | 8.000000 | 10.155000 |
| 75% | 10.000000 | 10.685000 |
| max | 15.000000 | 12.400000 |
Displays the number of C and T for f, respectively.
print('num of C:', len(df[df['f'] == 'C']))
print('num of T:', len(df[df['f'] == 'T']))
num of C: 50
num of T: 50
Descriptive statistics of the data confirm the following:
- Number of seeds y:
- Non-negative integers (2 to 15).
- Mean and variance are fairly close.
- Size of individuals x:
- Positive real numbers (continuous values)
- Variance is small.
- Fertilizer treatment f:
- It is a categorical variable.
- The number of elements in each category is equal.
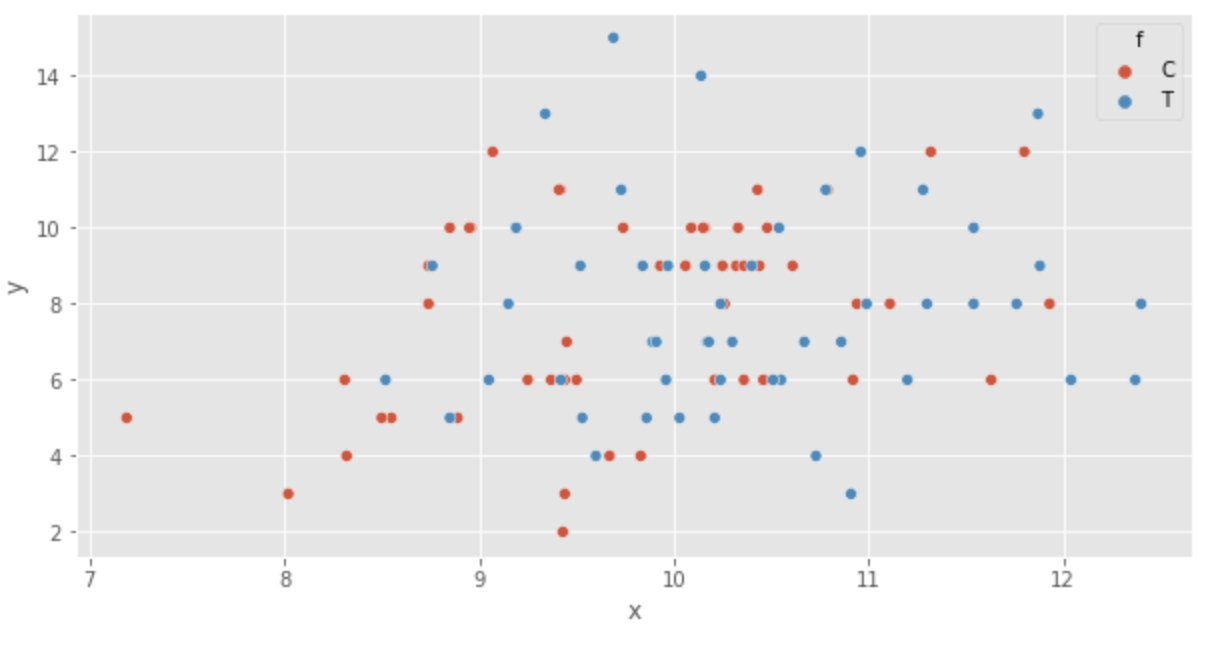
Next, we visualize the data. First, a scatter plot is displayed.
import matplotlib.pyplot as plt
import seaborn as sns
%matplotlib inline
plt.style.use('ggplot')
plt.figure(figsize=(10,5))
sns.set(rc={'figure.figsize':(10,5)})
sns.scatterplot(x='x', y='y', hue='f', data=df)
Displays a box-and-whisker diagram.
sns.boxplot(x='f', y='y', data=df)
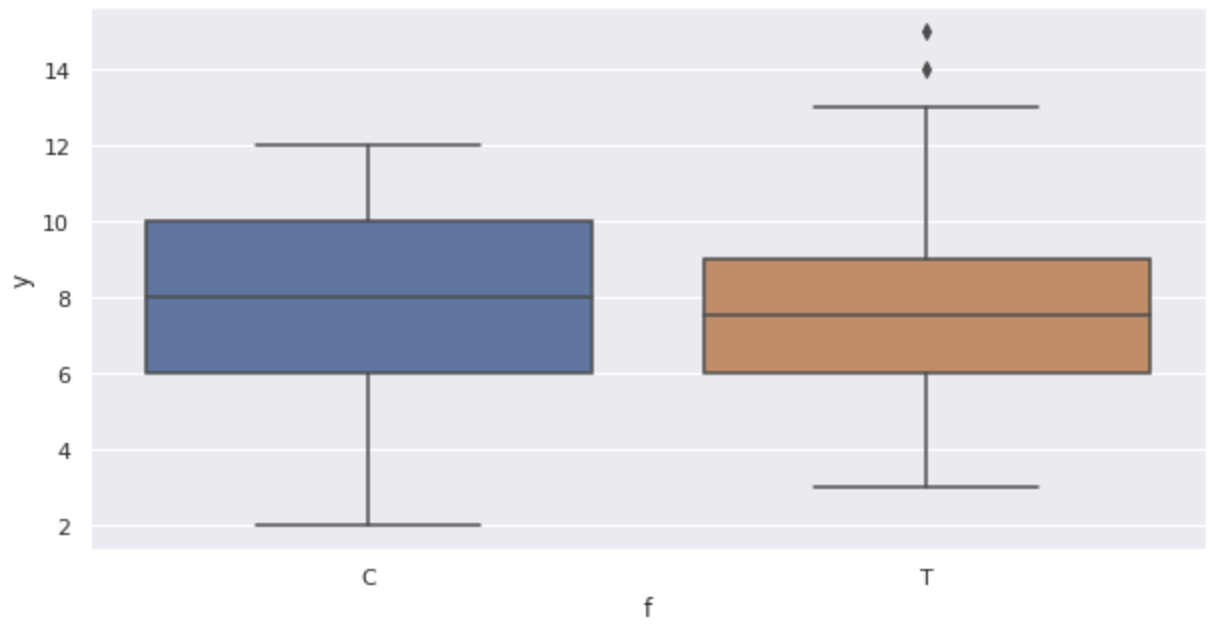
The graph confirms the following:
- As size
x - There appears to be no effect of fertilizer (box-and-whisker diagram)
Building a statistical model with Poisson regression
Assume that the number of seeds
The Poisson distribution has a parameter
- Statistical model where number of seeds depends on the size of the individual
- Statistical model where number of seeds depends on the presence or absence of fertilizer treatment
- Statistical model where number of seeds depends on the size of the individual and the presence or absence of fertilizer treatment.
Statistical model where number of seeds depends on the size of the individual
Let us first define
where
Taking the logarithm of both sides, we get
The right-hand side
The function on the left-hand side for the linear predictor is called the link function. This time the link function is
The log link function is often used in the GLM for Poisson regression. The reason is that the mean
From this we estimate
Parameter estimation is easily accomplished with the following Python code.
import statsmodels.api as sm
import statsmodels.formula.api as smf
fit_x = smf.glm('y ~ x', data=df, family=sm.families.Poisson()).fit()
fit_x.summary()
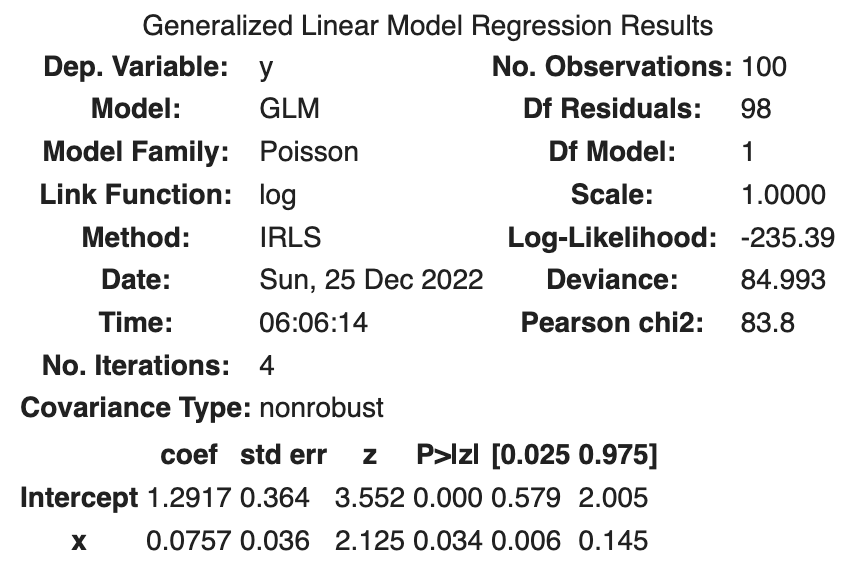
The following results were obtained.
| coef | std err | z | Log-likelihood | |
|---|---|---|---|---|
| Intercept | 1.2917 | 0.364 | 3.552 | |
| x | 0.0757 | 0.036 | 2.125 | |
| -235.39 |
std err is the standard error, i.e., the standard deviation of the estimated parameters. A normal distribution is assumed for the derivation of the standard deviation. z is the z-value.
The
Plot the above function.
import numpy as np
import math
sns.scatterplot(x='x', y='y', hue='f', data=df)
x = np.arange(df.x.min(), df.x.max() + 0.01, 0.01)
y = [math.exp(fit_x.params['Intercept'] + fit_x.params['x'] * x_i) for x_i in x]
plt.plot(x, y)
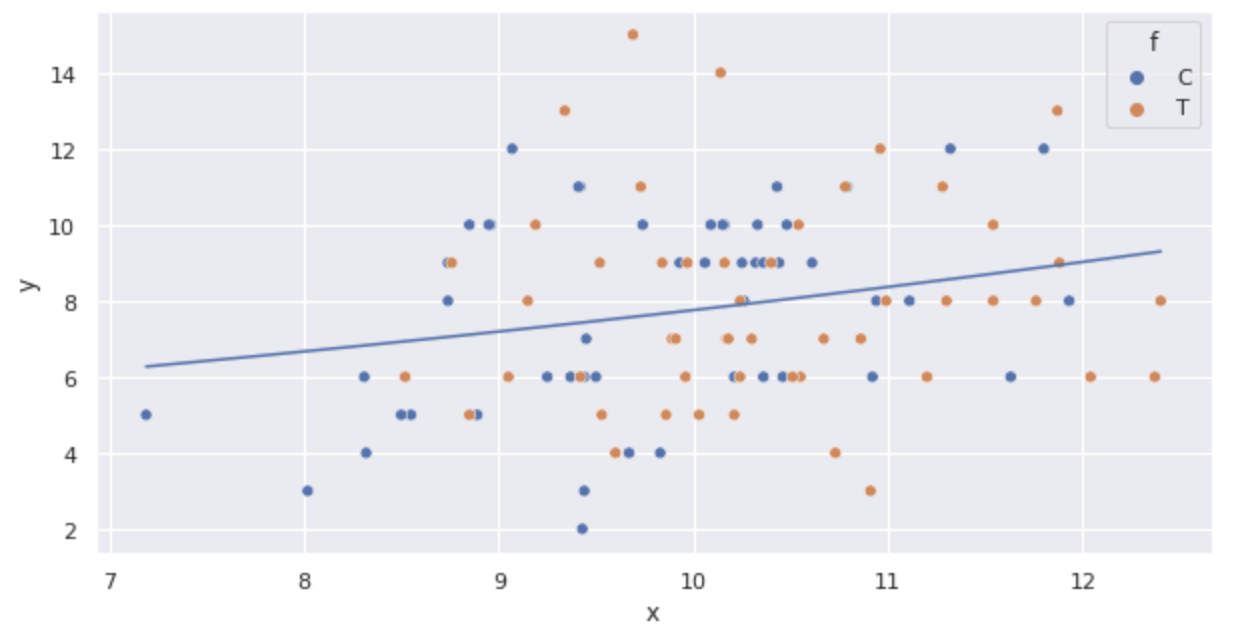
The line is drawn that the average seed count increases as size
Statistical model where number of seeds depends on the presence or absence of fertilizer treatment
Let us consider a model that incorporates fertilizer application status
Fertilizer application
The model is as follows.
That is, if individual
and if individual
Let us estimate
import statsmodels.api as sm
import statsmodels.formula.api as smf
fit_f = smf.glm('y ~ f', data=df, family=sm.families.Poisson()).fit()
fit_f.summary()
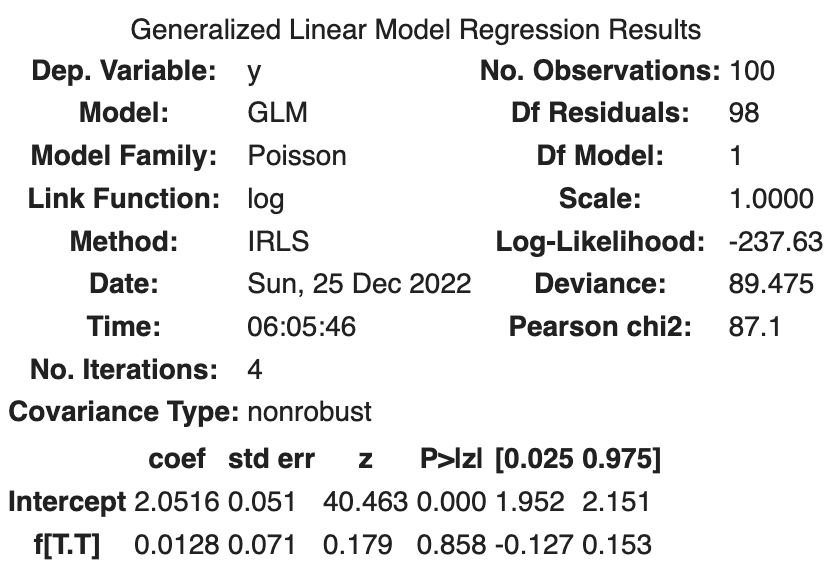
The following results were obtained.
| coef | std err | z | Log-likelihood | |
|---|---|---|---|---|
| Intercept | 2.0516 | 0.051 | 40.463 | |
| f[T.T] | 0.0128 | 0.071 | 0.179 | |
| -237.63 |
That is,
The addition of the fertilizer treatment resulted in a slight increase in the average number of seeds.
The log-likelihood is -237.63, which is smaller than the log-likelihood of the model with
Statistical model where number of seeds depends on the size of the individual and the presence or absence of fertilizer treatment
Finally, we consider a model that incorporates size
The model is as follows
Estimate
import statsmodels.api as sm
import statsmodels.formula.api as smf
fit_full = smf.glm('y ~ x + f', data=df, family=sm.families.Poisson()).fit()
fit_full.summary()
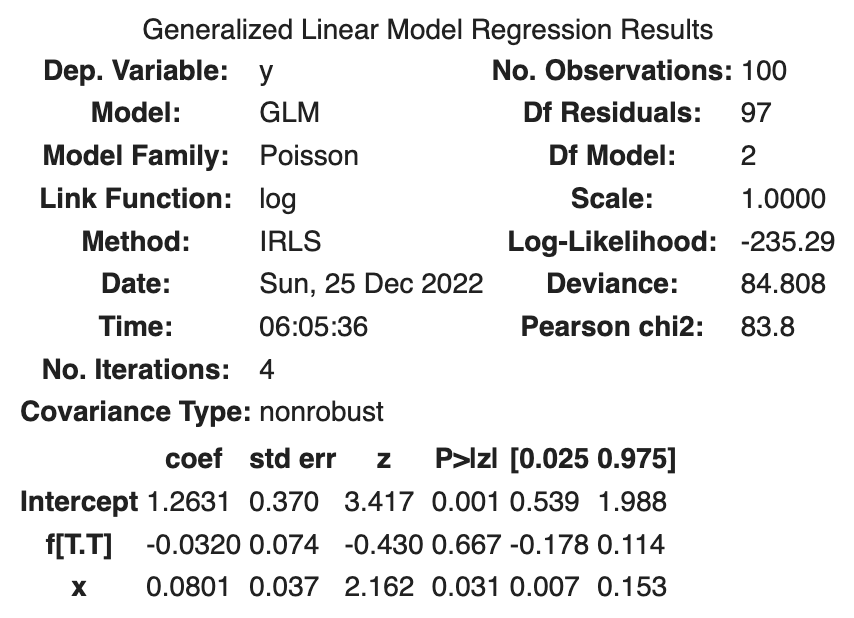
The following results were obtained.
| coef | std err | z | Log-likelihood | |
|---|---|---|---|---|
| Intercept | 1.2631 | 0.370 | 3.417 | |
| f[T.T] | -0.0320 | 0.074 | -0.430 | |
| x | 0.0801 | 0.037 | 2.162 | |
| -235.29 |
The following results were obtained.
It is interpreted that the average seed count will be lower with fertilizer treatment. Specifically,
The log-likelihood is -235.29, which is a better fit to the data than the previous two models.