What is logistic regression
Logistic regression is a GLM used to express observed events in terms of a binomial distribution.
The link function of the binomial distribution is often the logit link function, which corresponds to the left side of the following equation.
where
Building a logistic regression model
Let us actually construct a logistic regression model. Here we consider seed data for 100 fictitious plants. The data consists of the following columns:
- N: Number of seeds
- y: Number of germinated seeds
- x: Size of the individuals
- f: Whether the individuals were treated with fertilizer or not (C: not treated, T: treated)
import pandas as pd
# number of seeds
N = [8,8,8,8,8,8,8,8,8,8,8,8,8,8,8,8,8,8,8,8,8,8,8,8,8,8,8,8,8,8,8,8,8,8,8,8,8,8,8,
8,8,8,8,8,8,8,8,8,8,8,8,8,8,8,8,8,8,8,8,8,8,8,8,8,8,8,8,8,8,8,8,8,8,8,8,8,8,8,
8,8,8,8,8,8,8,8,8,8,8,8,8,8,8,8,8,8,8,8,8,8]
# number of seeds germinated
y = [1,6,5,6,1,1,3,6,0,8,0,2,0,5,3,6,3,4,5,8,5,8,1,4,1,8,4,7,5,5,8,1,7,1,3,8,6,7,
7,4,7,8,8,0,6,5,8,3,8,8,0,5,5,8,3,2,7,8,3,5,8,3,6,7,8,6,5,5,0,8,8,0,6,1,8,8,
6,6,6,6,8,8,7,8,6,2,0,7,8,8,8,3,7,8,8,7,0,5,8,1]
# body size
x = [9.76,10.48,10.83,10.94,9.37,8.81,9.49,11.02,7.97,11.55,9.46,9.47,8.71,10.42,10.06,
11,9.95,9.52,10.26,11.33,9.77,10.59,9.35,10,9.53,12.06,9.68,11.32,10.48,10.37,11.33,
9.42,10.68,7.91,9.39,11.65,10.66,11.23,10.57,10.42,11.73,12.02,11.55,8.58,11.08,10.49,
11.12,8.99,10.08,10.8,7.83,8.88,9.74,9.98,8.46,7.96,9.78,11.93,9.04,10.14,11.01,8.88,
9.68,9.8,10.76,9.81,8.37,9.38,7.68,10.23,9.83,7.66,9.33,8.2,9.54,10.55,9.88,9.34,10.38,
9.63,12.44,10.17,9.29,11.17,9.13,8.79,8.19,10.25,11.3,10.84,10.97,8.6,9.91,11.38,10.39,
10.45,8.94,8.94,10.14,8.5]
# feed (C: control, T: treatment)
f = ['C','C','C','C','C','C','C','C','C','C','C','C','C','C','C','C','C','C','C','C','C','C',
'C','C','C','C','C','C','C','C','C','C','C','C','C','C','C','C','C','C','C','C','C','C',
'C','C','C','C','C','C','T','T','T','T','T','T','T','T','T','T','T','T','T','T','T','T',
'T','T','T','T','T','T','T','T','T','T','T','T','T','T','T','T','T','T','T','T','T','T',
'T','T','T','T','T','T','T','T','T','T','T','T']
df = pd.DataFrame(list(zip(N, y, x, f)), columns =['N', 'y', 'x', 'f'])
display(df.head())
| N | y | x | f | |
|---|---|---|---|---|
| 0 | 8 | 1 | 9.76 | C |
| 1 | 8 | 6 | 10.48 | C |
| 2 | 8 | 5 | 10.83 | C |
| 3 | 8 | 6 | 10.94 | C |
| 4 | 8 | 1 | 9.37 | C |
Data investigation
Let us check descriptive statistics.
df.describe()
| N | y | x | |
|---|---|---|---|
| count | 100.0 | 100.000000 | 100.000000 |
| mean | 8.0 | 5.080000 | 9.967200 |
| std | 0.0 | 2.743882 | 1.088954 |
| min | 8.0 | 0.000000 | 7.660000 |
| 25% | 8.0 | 3.000000 | 9.337500 |
| 50% | 8.0 | 6.000000 | 9.965000 |
| 75% | 8.0 | 8.000000 | 10.770000 |
| max | 8.0 | 8.000000 | 12.440000 |
Displays the number of C and T for f, respectively.
print('num of C:', len(df[df['f'] == 'C']))
print('num of T:', len(df[df['f'] == 'T']))
num of C: 50
num of T: 50
The descriptive statistics of the data confirm the following
- Number of seeds N:
- All the numbers are in 8.
- Number of germinated seeds y:
- Non-negative integer (0 to 8)
- Size of individuals x:
- Positive real numbers (continuous values)
- Variance is small.
- Fertilizer treatment f:
- It is categorical data.
- The number of elements in each category is equal.
import matplotlib.pyplot as plt
import seaborn as sns
%matplotlib inline
plt.style.use('ggplot')
plt.figure(figsize=(10,5))
sns.set(rc={'figure.figsize':(10,5)})
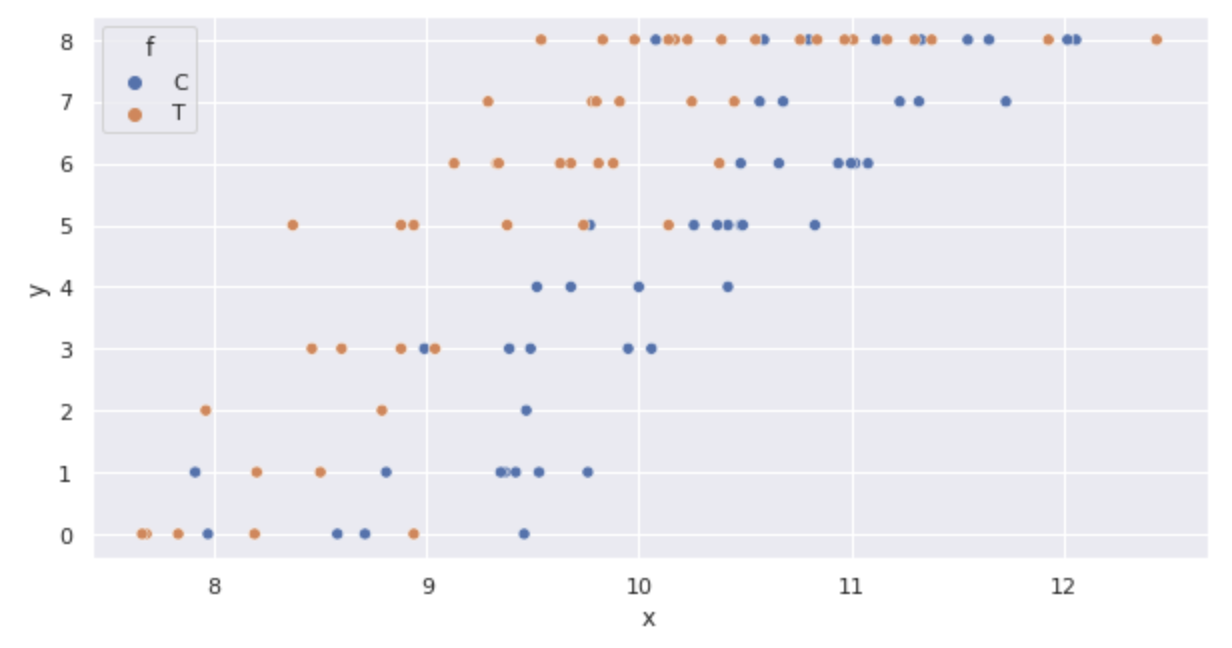
sns.scatterplot(x='x', y='y', hue='f', data=df)
Displays a box-and-whisker diagram.
sns.boxplot(x='f', y='y', data=df)
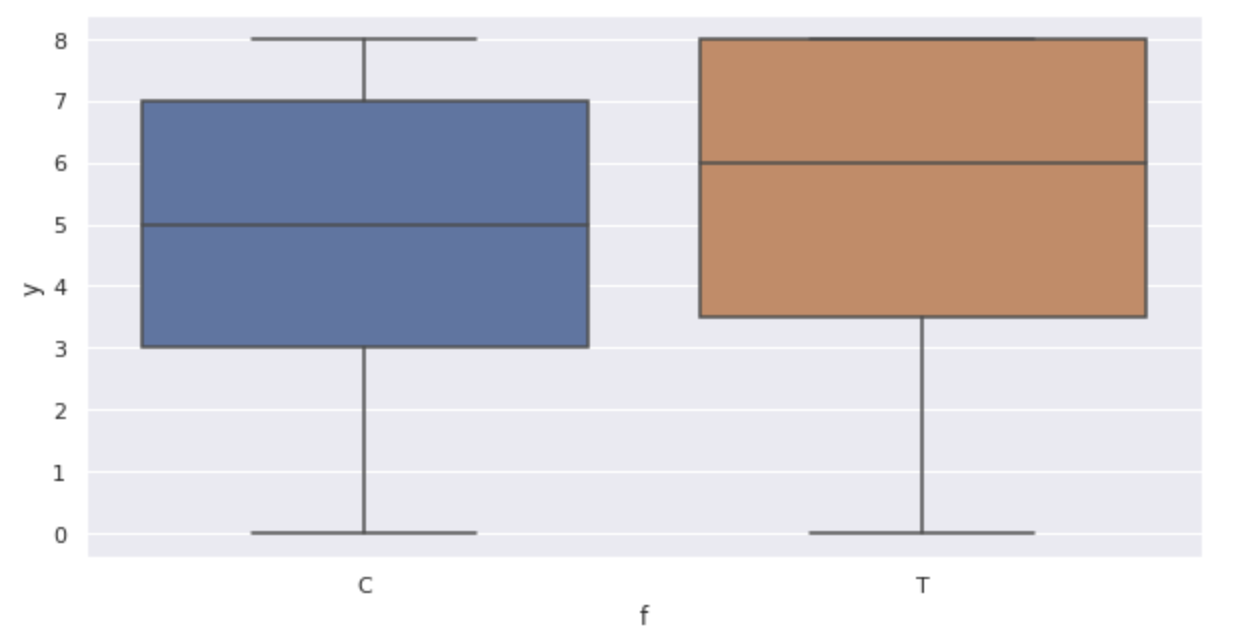
The graph confirms the following:
- As
x - The number of germinated seeds y also increases with fertilization (box-and-whisker plot)
Model Building
For
The binomial distribution is represented by the following equation:
Let
We consider a statistical model in which the germination probability
A function often used to express probability is the logistic function. The logistic function is expressed by the following equation:
The logistic function is illustrated below.
import matplotlib.pyplot as plt
import pandas as pd
import seaborn as sns
%matplotlib inline
plt.style.use('ggplot')
plt.figure(figsize=(10,5))
x = np.arange(-10, 10, 0.01)
y = [ 1 / ( 1 + math.exp(-x_i)) for x_i in x]
plt.plot(x, y, color='black', label='logsitic function')
plt.xlabel('x')
plt.ylabel('y')
plt.legend()
plt.show()
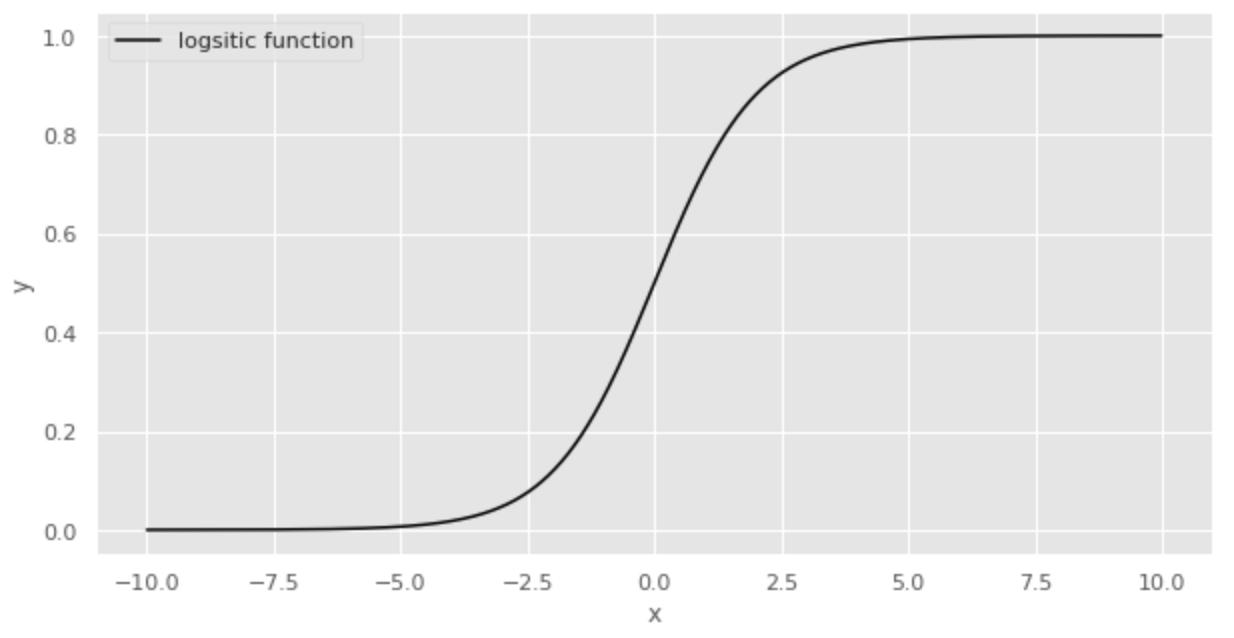
The logistic function ranges from 0 to 1 for y and 0.5 when x is 0. This property makes the logistic function a suitable function for expressing probability.
If we replace the logistic function with the variables in this case, we obtain the following equation.
where
The transformation for
The left-hand side of the above equation is called the logit link function. The logit link function is the inverse function of the logistic function.
Expressing the seed germination probability
That is, the germination probability
Therefore, we are looking for
Parameter estimation
The
import statsmodels.api as sm
import statsmodels.formula.api as smf
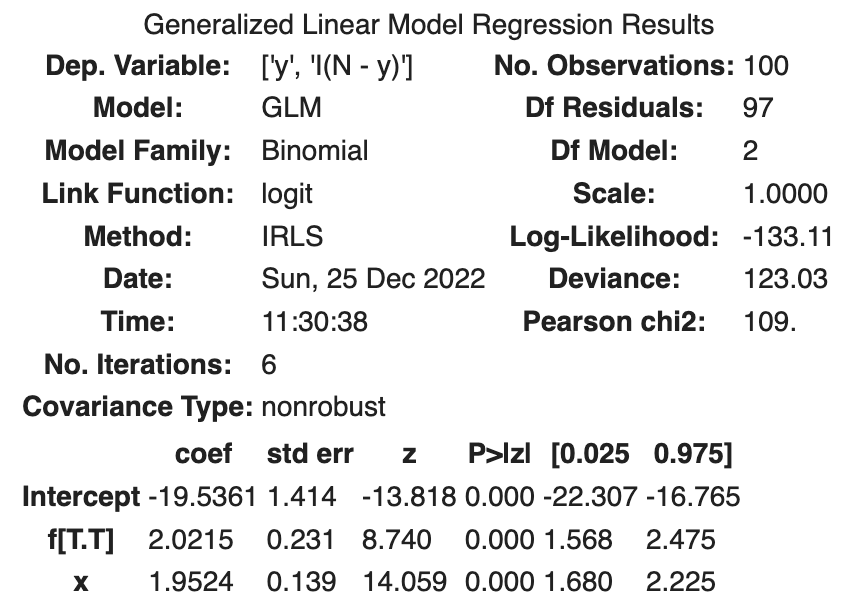
fit_xf = smf.glm(formula='y + I(N - y) ~ x + f', data=df, family=sm.families.Binomial()).fit()
fit_xf.summary()
# aic
fit_xf.aic
The following results were obtained
| coef | std err | z | Log-likelihood | AIC | |
|---|---|---|---|---|---|
| Intercept | -19.5361 | 1.414 | -13.818 | ||
| f[T.T] | 2.0215 | 0.231 | 8.740 | ||
| x | 1.9524 | 0.139 | 14.059 | ||
| -133.11 | 272.21 |
Substitute the estimated parameters into the logistic regression model and illustrate.
import numpy as np
import math
sns.scatterplot(x='x', y='y', hue='f', data=df)
x = np.arange(df.x.min(), df.x.max() + 0.01, 0.01)
y_T = [8 * 1 / ( 1 + math.exp(-(fit_xf.params['Intercept'] + fit_xf.params['x'] * x_i + fit_xf.params['f[T.T]']))) for x_i in x]
y_C = [8 * 1 / ( 1 + math.exp(-(fit_xf.params['Intercept'] + fit_xf.params['x'] * x_i))) for x_i in x]
plt.plot(x, y_T, color='black', label='logsitic T')
plt.plot(x, y_C, color='gray', label='logsitic C')
plt.legend()
plt.show()
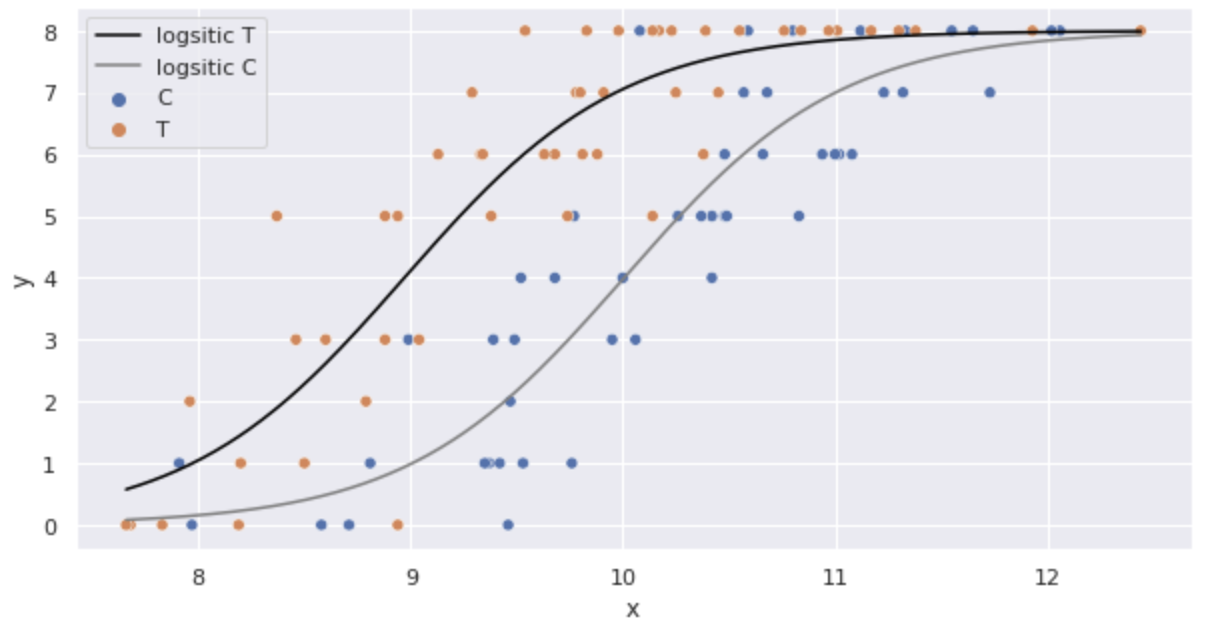
It looks like a good model is being built.
Interpreting logit
Let us transform the logit link function as follows:
The left side
The odds are proportional to
This means that the odds are 7.5 times greater with the fertilizer treatment compared to the no fertilizer treatment.
The logit link function is defined as