What is Generalized Linear Mixed Model (GLMM)
Generalized Linear Mixed Model (GLMM) is a statistical model that is a further development of GLM. It can represent unobservable individual and regional differences.
Unobservable individual and regional differences include, for example, the following:
- Individual differences in pain perception
Different people respond differently to the same pain. - Individual differences in scoring
Some scorers are more lenient than others. - Regional differences
There are regional variations in the nature of the pain.
A model that does not account for these individual and regional differences will lead to overdispersion. Overdispersion means that the assumed probability distribution has a larger variance than assumed. The following figure shows overdispersion using the Poisson distribution as an example.
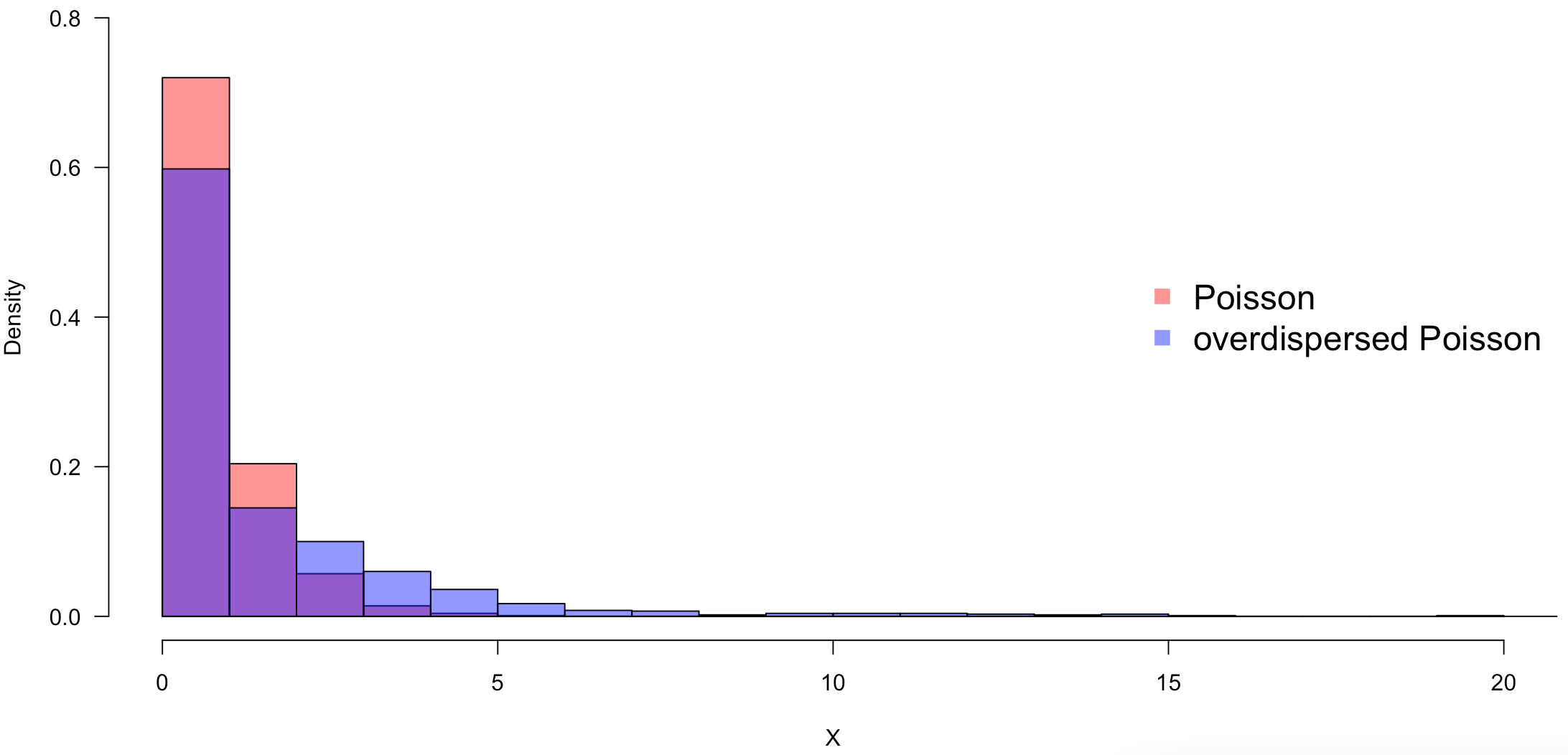
Individual and regional differences can result in a Poisson distribution that is larger than the original Poisson distribution, as in the case of overdispersed Poisson, and without taking individual and location differences into account, the observed data cannot be modeled well.
GLMM treats unexplained variation such as individual and location differences as random effects and models them as such, with determining that it is impossible to observe all of the explanatory variables.
Random Effects
GLMM adds random effects to linear predictors. For example, for individual
We assume that the random effect
Maximum likelihood estimation of the parameters will be done for
Cases where GLMM is required
The criterion for determining whether a GLMM is necessary is whether you are sampling the same individuals or locations over and over again. For example, GLMM is often applied to panel data, which are data observed repeatedly over time for individuals.
Build GLMM
Let us construct a GLMM using the dataset grouseticks. grouseticks is an observation of the number of ticks on the heads of red grouse chicks and consists of the following columns:
INDEX: (factor) chick number (observation level)TICKS: number of ticks sampledBROOD: (factor) brood numberHEIGHT: height above sea level (meters)YEAR: year (-1900)LOCATION: (factor) geographic location codecHEIGHT: centered height, derived from HEIGHT
Let us check the data.
> require(lme4)
> library(lme4)
> data(grouseticks)
> summary(grouseticks)
INDEX TICKS BROOD HEIGHT YEAR LOCATION cHEIGHT
1 : 1 Min. : 0.00 606 : 10 Min. :403.0 95:117 14 : 24 Min. :-59.241
2 : 1 1st Qu.: 0.00 602 : 9 1st Qu.:430.0 96:155 4 : 20 1st Qu.:-32.241
3 : 1 Median : 2.00 537 : 7 Median :457.0 97:131 19 : 20 Median : -5.241
4 : 1 Mean : 6.37 601 : 7 Mean :462.2 28 : 19 Mean : 0.000
5 : 1 3rd Qu.: 6.00 643 : 7 3rd Qu.:494.0 50 : 17 3rd Qu.: 31.759
6 : 1 Max. :85.00 711 : 7 Max. :533.0 36 : 16 Max. : 70.759
(Other):397 (Other):356 (Other):287
> head(grouseticks)
INDEX TICKS BROOD HEIGHT YEAR LOCATION cHEIGHT
1 1 0 501 465 95 32 2.759305
2 2 0 501 465 95 32 2.759305
3 3 0 502 472 95 36 9.759305
4 4 0 503 475 95 37 12.759305
5 5 0 503 475 95 37 12.759305
6 6 3 503 475 95 37 12.759305
> plot(TICKS ~ HEIGHT, las=1)
Poisson regression without random effects
This time we will construct a Poisson regression model. Using the average number of ticks
where Year.
The following code can be used to estimate the parameters.
> fit_glm <- glm(TICKS ~ YEAR, family=poisson(), data=grouseticks)
> summary)(fit_glm)
Call:
glm(formula = TICKS ~ YEAR, family = poisson(), data = grouseticks)
Deviance Residuals:
Min 1Q Median 3Q Max
-4.7110 -2.8888 -1.5183 0.7139 17.1467
Coefficients:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 1.78318 0.03790 47.05 <2e-16 ***
YEAR96 0.62348 0.04492 13.88 <2e-16 ***
YEAR97 -1.64109 0.08977 -18.28 <2e-16 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
(Dispersion parameter for poisson family taken to be 1)
Null deviance: 5847.5 on 402 degrees of freedom
Residual deviance: 4549.4 on 400 degrees of freedom
AIC: 5486.4
Number of Fisher Scoring iterations: 6
AIC is 5486.4.
Poisson regression with random effects
let us construct a Poisson regression model with random effects.
Random intercept model
Add the random effect LOCATION and express the average number of ticks
This model is also called the random intercept model because random effects are included in the intercept.
You can estimate TICKS ~ YEAR + (1 | LOCATION).
> fit_glmm <- glmer(TICKS ~ YEAR + (1 | LOCATION), family="poisson", data=grouseticks)
> summary(fit_glmm)
Generalized linear mixed model fit by maximum likelihood (Laplace Approximation) ['glmerMod']
Family: poisson ( log )
Formula: TICKS ~ YEAR + (1 | LOCATION)
Data: grouseticks
AIC BIC logLik deviance df.resid
2306.3 2322.3 -1149.1 2298.3 399
Scaled residuals:
Min 1Q Median 3Q Max
-6.6399 -0.8844 -0.4708 0.8543 5.5757
Random effects:
Groups Name Variance Std.Dev.
LOCATION (Intercept) 1.644 1.282
Number of obs: 403, groups: LOCATION, 63
Fixed effects:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 0.64992 0.18319 3.548 0.000388 ***
YEAR96 0.94371 0.08492 11.113 < 2e-16 ***
YEAR97 -1.43642 0.12298 -11.681 < 2e-16 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Correlation of Fixed Effects:
(Intr) YEAR96
YEAR96 -0.295
YEAR97 -0.241 0.520
The AIC is now 2306.3, a significant improvement over the model without random effects.
The variance of random effects for LOCATION is 1.644. You can check the random effects of all LOCATION with the randf() function.
> ranef(fit_glmm)
$LOCATION
(Intercept)
1 1.24539318
2 0.92293852
3 -0.93190888
.
.
.
61 -1.35144088
62 -0.95446024
63 -1.15277383
with conditional variances for “LOCATION”
The values from 1 to 63 are the LOCATION values: intercept for 1 is 1.24539318 and intercept for 3 is -0.93190888. The interpretation is that LOCATION 1 has relatively more ticks than LOCATION 3.
In addition, a random effect INDEX can be added.
> fit_glmm <- glmer(TICKS ~ YEAR + (1 | LOCATION) + (1 | INDEX), family="poisson", data=grouseticks)
> summary(fit_glmm)
Generalized linear mixed model fit by maximum likelihood (Laplace Approximation) ['glmerMod']
Family: poisson ( log )
Formula: TICKS ~ YEAR + (1 | LOCATION) + (1 | INDEX)
Data: grouseticks
AIC BIC logLik deviance df.resid
1863.9 1883.9 -927.0 1853.9 398
Scaled residuals:
Min 1Q Median 3Q Max
-1.5003 -0.5933 -0.1154 0.2723 2.3743
Random effects:
Groups Name Variance Std.Dev.
INDEX (Intercept) 0.5081 0.7128
LOCATION (Intercept) 1.4679 1.2116
Number of obs: 403, groups: INDEX, 403; LOCATION, 63
Fixed effects:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 0.2774 0.2107 1.316 0.188
YEAR96 1.2578 0.1821 6.906 4.97e-12 ***
YEAR97 -1.0856 0.2010 -5.400 6.67e-08 ***
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Correlation of Fixed Effects:
(Intr) YEAR96
YEAR96 -0.536
YEAR97 -0.439 0.552
The AIC is now 1863.9, which is a further improvement.
The variance of random effects for LOCATION is 1.4679 and for INDEX is 0.5081. We can see that the random effect of INDEX is smaller than that of LOCATION.
Random coefficient model
Let us add the random effect LOCATION not only to the intercept but also to the coefficient.
This model is also called the random coefficient model because random effects are included in the coefficients.
The following code can estimate TICKS ~ YEAR + (YEAR | LOCATION).
> fit_glmm <- glmer(TICKS ~ YEAR + (YEAR | LOCATION), family="poisson", data=grouseticks)
> summary(fit_glmm)
Generalized linear mixed model fit by maximum likelihood (Laplace Approximation) ['glmerMod']
Family: poisson ( log )
Formula: TICKS ~ YEAR + (YEAR | LOCATION)
Data: grouseticks
AIC BIC logLik deviance df.resid
2266.7 2302.7 -1124.3 2248.7 394
Scaled residuals:
Min 1Q Median 3Q Max
-6.6442 -0.8112 -0.4797 0.8437 5.5779
Random effects:
Groups Name Variance Std.Dev. Corr
LOCATION (Intercept) 2.3583 1.5357
YEAR96 0.6002 0.7747 -0.70
YEAR97 0.6735 0.8207 -0.80 0.46
Number of obs: 403, groups: LOCATION, 63
Fixed effects:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 0.3273 0.2674 1.224 0.22083
YEAR96 1.3094 0.2542 5.152 2.58e-07 ***
YEAR97 -0.7578 0.2909 -2.605 0.00918 **
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Correlation of Fixed Effects:
(Intr) YEAR96
YEAR96 -0.746
YEAR97 -0.668 0.553
However, this estimation is for a model that assumes the correlation between TICKS ~ YEAR + (1 | LOCATION) + (0 + YEAR | LOCATION).
> fit_glmm <- glmer(TICKS ~ YEAR + (1 | LOCATION) + (0 + YEAR | LOCATION), family="poisson", data=grouseticks)
> summary(fit_glmm)
Generalized linear mixed model fit by maximum likelihood (Laplace Approximation) ['glmerMod']
Family: poisson ( log )
Formula: TICKS ~ YEAR + (1 | LOCATION) + (0 + YEAR | LOCATION)
Data: grouseticks
AIC BIC logLik deviance df.resid
2268.7 2308.7 -1124.3 2248.7 393
Scaled residuals:
Min 1Q Median 3Q Max
-6.6442 -0.8112 -0.4797 0.8437 5.5779
Random effects:
Groups Name Variance Std.Dev. Corr
LOCATION (Intercept) 0.4151 0.6443
LOCATION.1 YEAR95 1.9431 1.3939
YEAR96 0.8755 0.9357 0.85
YEAR97 0.6006 0.7750 0.87 0.54
Number of obs: 403, groups: LOCATION, 63
Fixed effects:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 0.3274 0.2674 1.224 0.22081
YEAR96 1.3094 0.2542 5.152 2.58e-07 ***
YEAR97 -0.7578 0.2909 -2.605 0.00918 **
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Correlation of Fixed Effects:
(Intr) YEAR96
YEAR96 -0.746
YEAR97 -0.668 0.553
It is also possible to add random effects only to coefficients by writing TICKS ~ YEAR + (0 + YEAR | LOCATION).
> fit_glmm <- glmer(TICKS ~ YEAR + (0 + YEAR | LOCATION), family="poisson", data=grouseticks)
Warning message:
In checkConv(attr(opt, "derivs"), opt$par, ctrl = control$checkConv, :
Model failed to converge with max|grad| = 0.0139508 (tol = 0.002, component 1)
> summary(fit_glmm)
Generalized linear mixed model fit by maximum likelihood (Laplace Approximation) ['glmerMod']
Family: poisson ( log )
Formula: TICKS ~ YEAR + (0 + YEAR | LOCATION)
Data: grouseticks
AIC BIC logLik deviance df.resid
2266.7 2302.7 -1124.3 2248.7 394
Scaled residuals:
Min 1Q Median 3Q Max
-6.6442 -0.8114 -0.4793 0.8432 5.5778
Random effects:
Groups Name Variance Std.Dev. Corr
LOCATION YEAR95 2.363 1.537
YEAR96 1.291 1.136 0.87
YEAR97 1.016 1.008 0.87 0.70
Number of obs: 403, groups: LOCATION, 63
Fixed effects:
Estimate Std. Error z value Pr(>|z|)
(Intercept) 0.3276 0.2676 1.224 0.22093
YEAR96 1.3099 0.2543 5.151 2.59e-07 ***
YEAR97 -0.7566 0.2908 -2.601 0.00929 **
---
Signif. codes: 0 ‘***’ 0.001 ‘**’ 0.01 ‘*’ 0.05 ‘.’ 0.1 ‘ ’ 1
Correlation of Fixed Effects:
(Intr) YEAR96
YEAR96 -0.746
YEAR97 -0.669 0.554
optimizer (Nelder_Mead) convergence code: 0 (OK)
Model failed to converge with max|grad| = 0.0139508 (tol = 0.002, component 1)
R code
The R code to illustrate the Poisson distribution overdispersion is shown below.
> set.seed(1)
> hist(Y1 <- rpois(1000, 1), breaks=seq(0, 20), col="#FF00007F", freq=F, ylim=c(0, 0.8), las=1,
main="", xlab="X")
> hist(Y2 <- rpois(1000, 1 * exp(rnorm(1000, mean=0, sd=1))), col="#0000FF7F", add=T, freq=F, breaks=seq(0, 100))
> legend("right", legend=c("Poisson", "overdispersed Poisson"), pch=15, col=c("#FF00007F", "#0000FF7F"),
bty="n", cex=1.5)
