What is PCA
Principal Component Analysis (PCA) is a widely used technique in the realm of data science, particularly for dimensionality reduction, data visualization, and noise reduction. This powerful method enables researchers and practitioners to analyze and uncover the hidden patterns within large and complex datasets, which can lead to valuable insights and improved decision-making.
The Essence of PCA
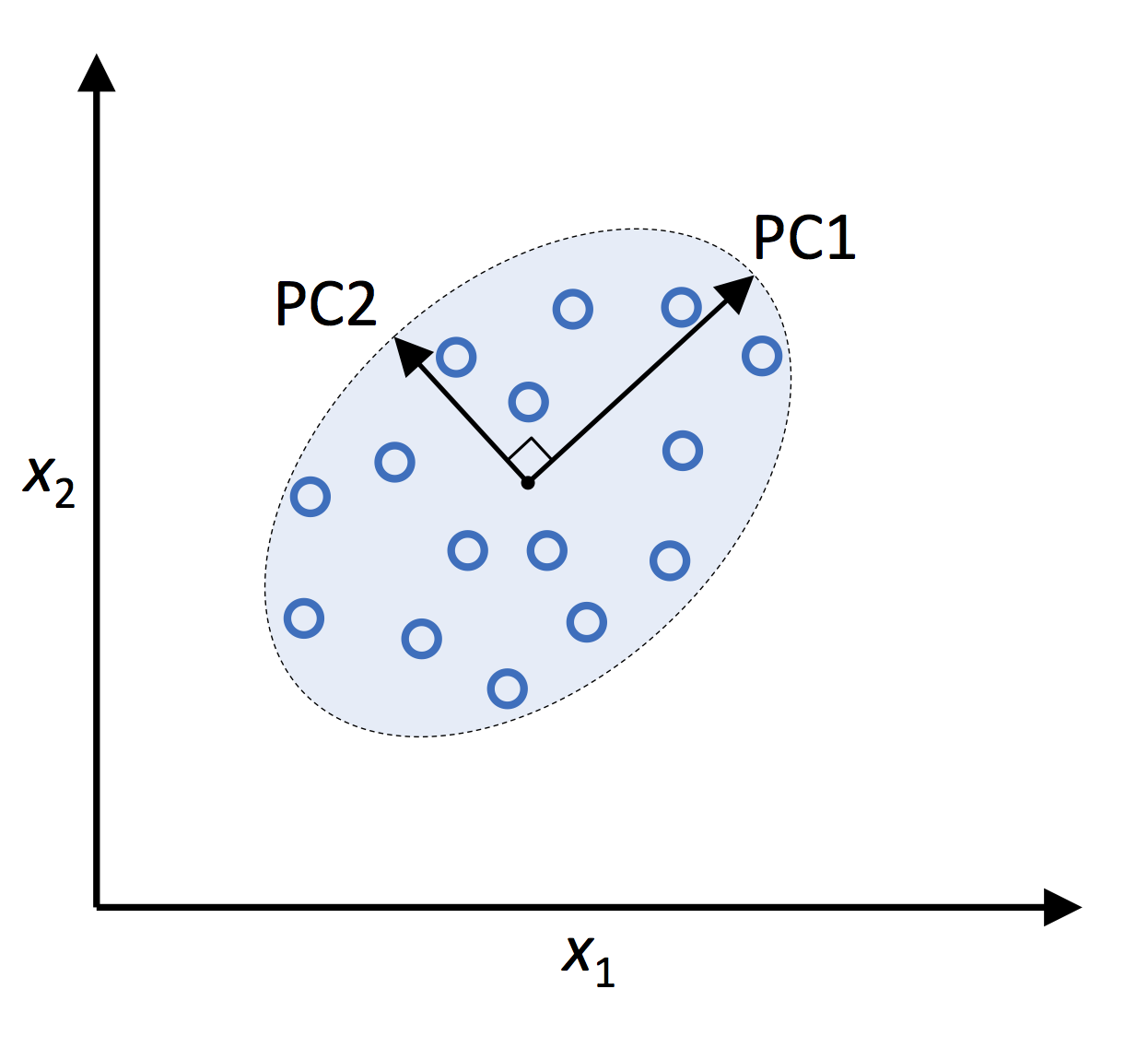
The primary goal of PCA is to reduce the dimensionality of a dataset while retaining as much information as possible. This is achieved by transforming the original features into a new set of orthogonal features, called principal components (PCs), which are linear combinations of the original variables. The PCs are ranked in order of their variance, with the first principal component accounting for the most variance in the data, followed by the second, and so on. By retaining only a subset of PCs, one can effectively reduce the dimensionality of the dataset and simplify analysis without losing significant information.
Python Machine Learning (2nd Ed.) Code Repository
Mathematics of PCA
In this chapter, I delve into the mathematical foundations of PCA. To understand the underlying concepts and techniques, a basic knowledge of linear algebra and statistics is essential.
Linear Algebra and Eigenvalues
PCA relies on linear algebra concepts, specifically eigenvalues and eigenvectors. Given a square matrix
where
Covariance Matrix
The covariance matrix is a key element in PCA, as it captures the relationships between the original variables in the dataset. For a dataset
where
Eigendecomposition
Eigendecomposition is the process of decomposing a matrix into its eigenvalues and eigenvectors. In PCA, we perform eigendecomposition on the covariance matrix
Dimensionality Reduction
Once we have the eigenvalues and eigenvectors of the covariance matrix, we can reduce the dimensionality of the dataset by selecting the top
where
PCA in Practice
In this chapter, I will walk through a step-by-step guide on performing PCA using a publicly available dataset - the famous Iris dataset. This dataset contains 150 samples of iris flowers, with four features: sepal length, sepal width, petal length, and petal width. The objective is to reduce the dimensionality of the dataset while preserving its inherent structure.
Data Preprocessing
Before performing PCA, it is essential to preprocess the data. The main preprocessing step is standardization. Standardization scales the features to have a mean of 0 and a standard deviation of 1. This step is necessary for PCA to work effectively.
Here's how to preprocess the Iris dataset using Python and the scikit-learn library:
import numpy as np
import pandas as pd
from sklearn import datasets
from sklearn.preprocessing import StandardScaler
# Load the Iris dataset
iris = datasets.load_iris()
X = iris.data
# Standardize the data
scaler = StandardScaler()
X_scaled = scaler.fit_transform(X)
Implementing PCA
Now that the data is preprocessed, we can perform PCA using the scikit-learn library. Here's the code to reduce the Iris dataset to two dimensions:
from sklearn.decomposition import PCA
# Perform PCA with 2 components
pca = PCA(n_components=2)
X_pca = pca.fit_transform(X_scaled)
Interpretation of PCA Results
The output of PCA is a new dataset (X_pca) with reduced dimensions. In our example, we've reduced the Iris dataset from four dimensions to two. To understand the importance of each principal component, we can examine the explained variance ratio, which indicates the proportion of the total variance in the data explained by each component:
explained_variance_ratio = pca.explained_variance_ratio_
print(explained_variance_ratio)
The output might be something like:
array([0.72962445, 0.22850762])
This indicates that the first principal component explains approximately 72.96% of the total variance in the data, while the second principal component explains about 22.85%. Together, these two components account for roughly 95.81% of the total variance.
Visualizing PCA Components
Finally, we can visualize the PCA-transformed data in a scatter plot. This will allow us to observe any patterns or clusters in the reduced-dimensionality dataset. Here's the code to create a scatter plot of the two principal components using the matplotlib:
import matplotlib.pyplot as plt
# Plot the PCA-transformed data
plt.scatter(X_pca[:, 0], X_pca[:, 1], c=iris.target, cmap='viridis', edgecolor='k', s=75)
plt.xlabel('First Principal Component')
plt.ylabel('Second Principal Component')
plt.title('PCA with 2 Components on Iris Dataset')
plt.show()
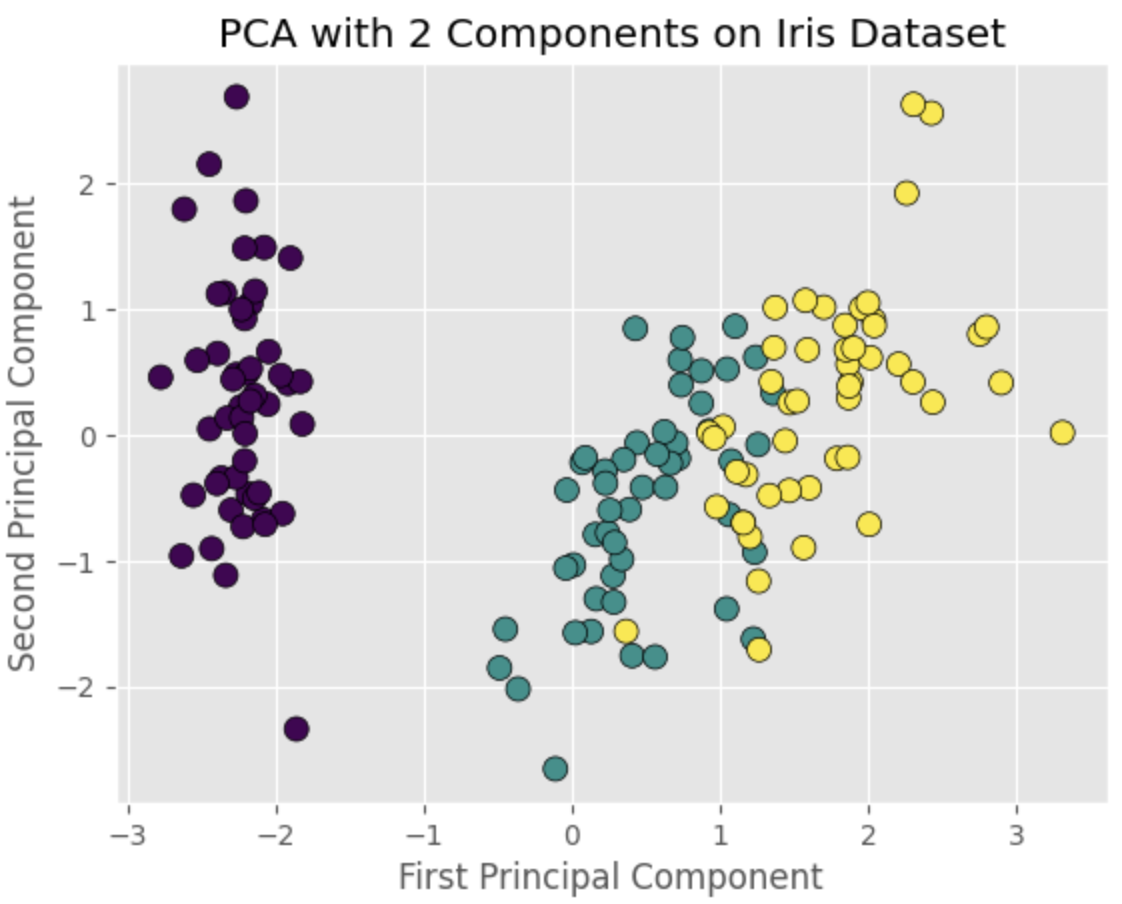
The scatter plot will show distinct clusters corresponding to the three iris species, demonstrating that PCA can effectively reduce the dimensionality of the data while preserving its structure.
Principal Component Loadings
Principal component loadings, also known as component loadings or eigenvector loadings, are the coefficients that indicate how each original feature contributes to a particular principal component. Analyzing the loadings can help us understand the relationships between the original features and the principal components, as well as the underlying structure of the data.
Here's how to obtain the principal component loadings for the Iris dataset using the PCA model we implemented earlier:
loadings = pca.components_
print(loadings)
The output might be something like:
array([[ 0.52106591, -0.26934744, 0.5804131 , 0.56485654],
[ 0.37741762, 0.92329566, 0.02449161, 0.06694199]])
This array represents the loadings for the two principal components, with each row corresponding to a principal component and each column corresponding to an original feature. The higher the absolute value of a loading, the stronger the contribution of the corresponding original feature to the principal component.
For instance, in the first principal component, petal length (0.580) and petal width (0.565) have higher loadings compared to sepal length (0.521) and sepal width (-0.269). This suggests that the first principal component is largely driven by the variation in petal length and petal width.
To visualize the loadings, we can create a biplot, which is a scatter plot of the PCA-transformed data overlaid with vectors representing the loadings:
def biplot(X_pca, loadings, labels=None):
plt.figure(figsize=(10, 7))
plt.scatter(X_pca[:, 0], X_pca[:, 1], c=iris.target, cmap='viridis', edgecolor='k', s=75)
if labels is None:
labels = np.arange(loadings.shape[1])
for i, label in enumerate(labels):
plt.arrow(0, 0, loadings[0, i], loadings[1, i], color='r', alpha=0.8, lw=2)
plt.text(loadings[0, i] * 1.15, loadings[1, i] * 1.15, label, color='r', ha='center', va='center', fontsize=12)
plt.xlabel('First Principal Component')
plt.ylabel('Second Principal Component')
plt.title('PCA Biplot of Iris Dataset')
plt.grid()
plt.show()
feature_labels = iris.feature_names
biplot(X_pca, loadings, labels=feature_labels)
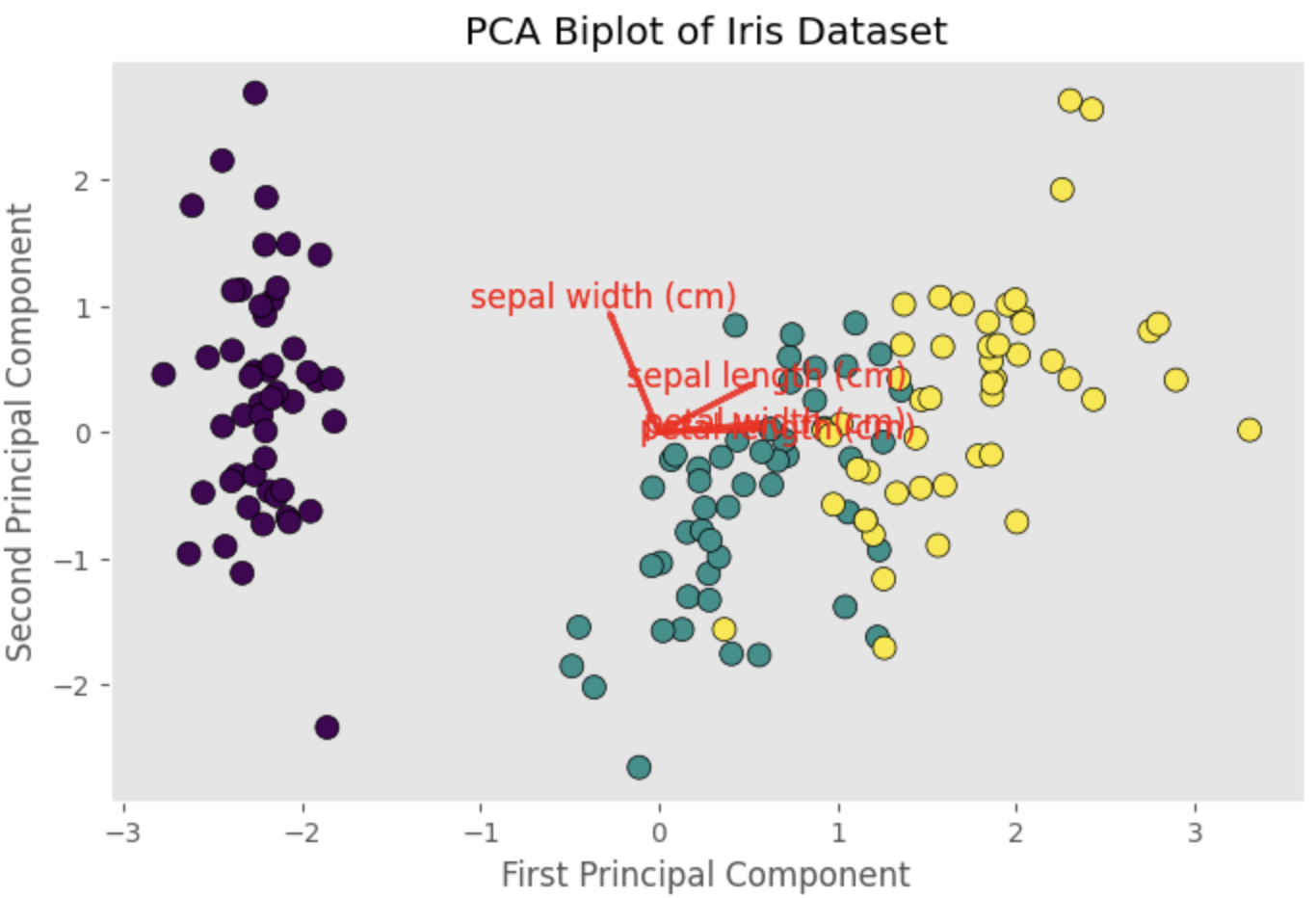
The biplot will display vectors representing the loadings for sepal length, sepal width, petal length, and petal width. The direction and length of each vector indicate the contribution of the corresponding feature to the principal components. By examining the biplot, we can gain insights into the relationships between the original features and the principal components, as well as the structure of the data.
References